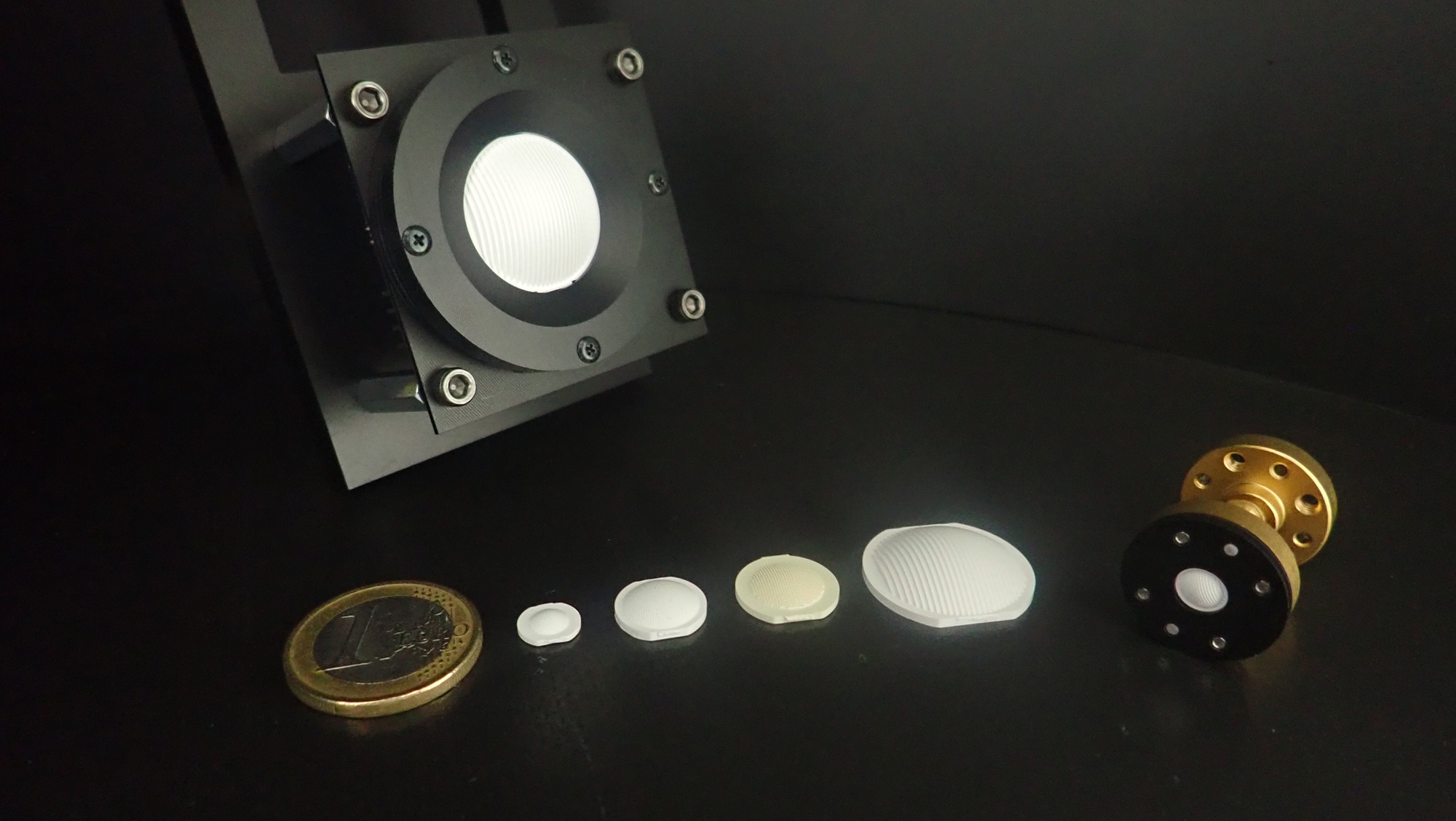
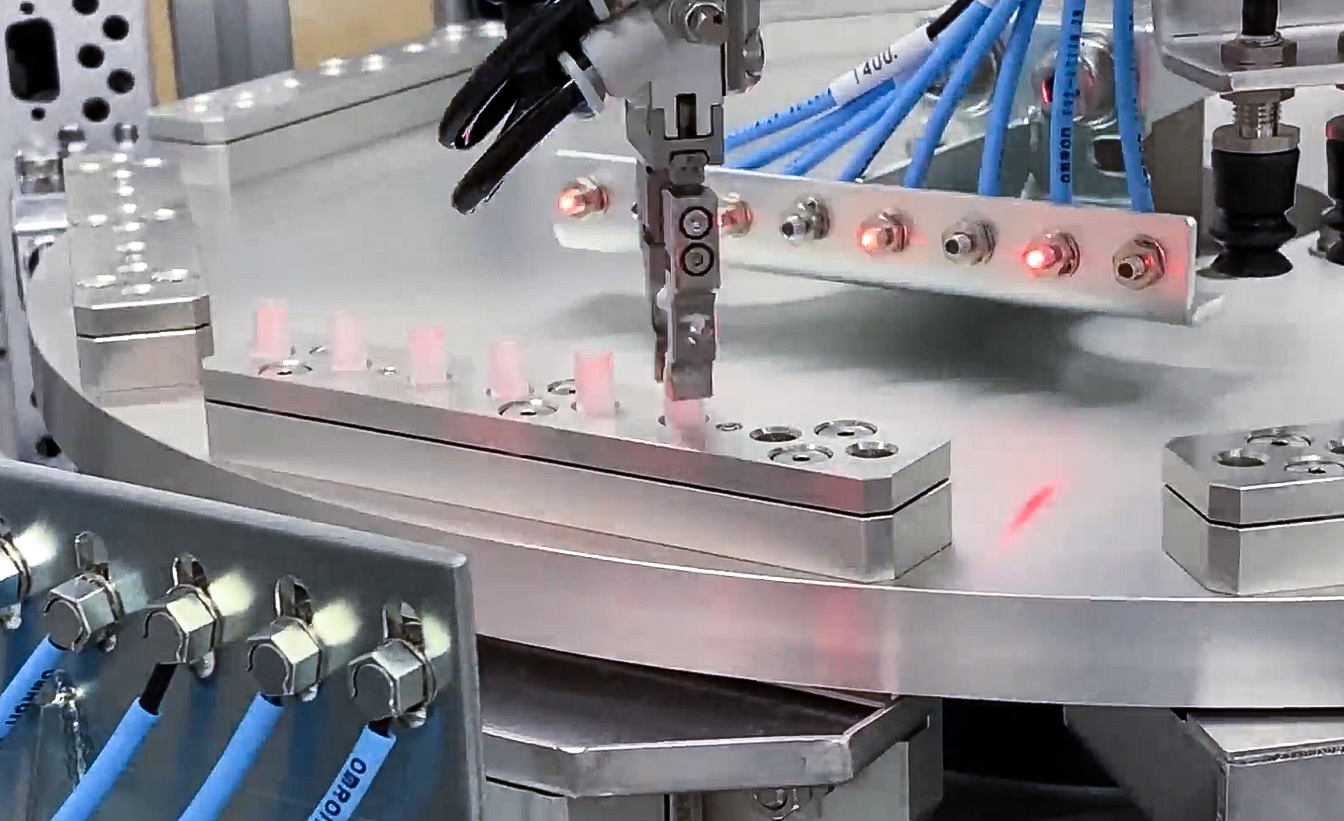
Performance-Focused Manufacturing Solutions.
Customer-Centric Plastics Engineering.
Quality-Focused Delivery.
Transform your innovative designs into high-volume, reliable commercial product delivery.
Access World-Class Engineering and Manufacturing of Custom Plastic Products for:
Optical, mechanical, microfluidic, or other high-performance requirements
Successful and innovative product development
Reliable high-volume product delivery
Custom Plastic Solutions for Demanding Industries
Custom plastic devices and parts engineered and manufactured by Enplas power demanding industries worldwide.

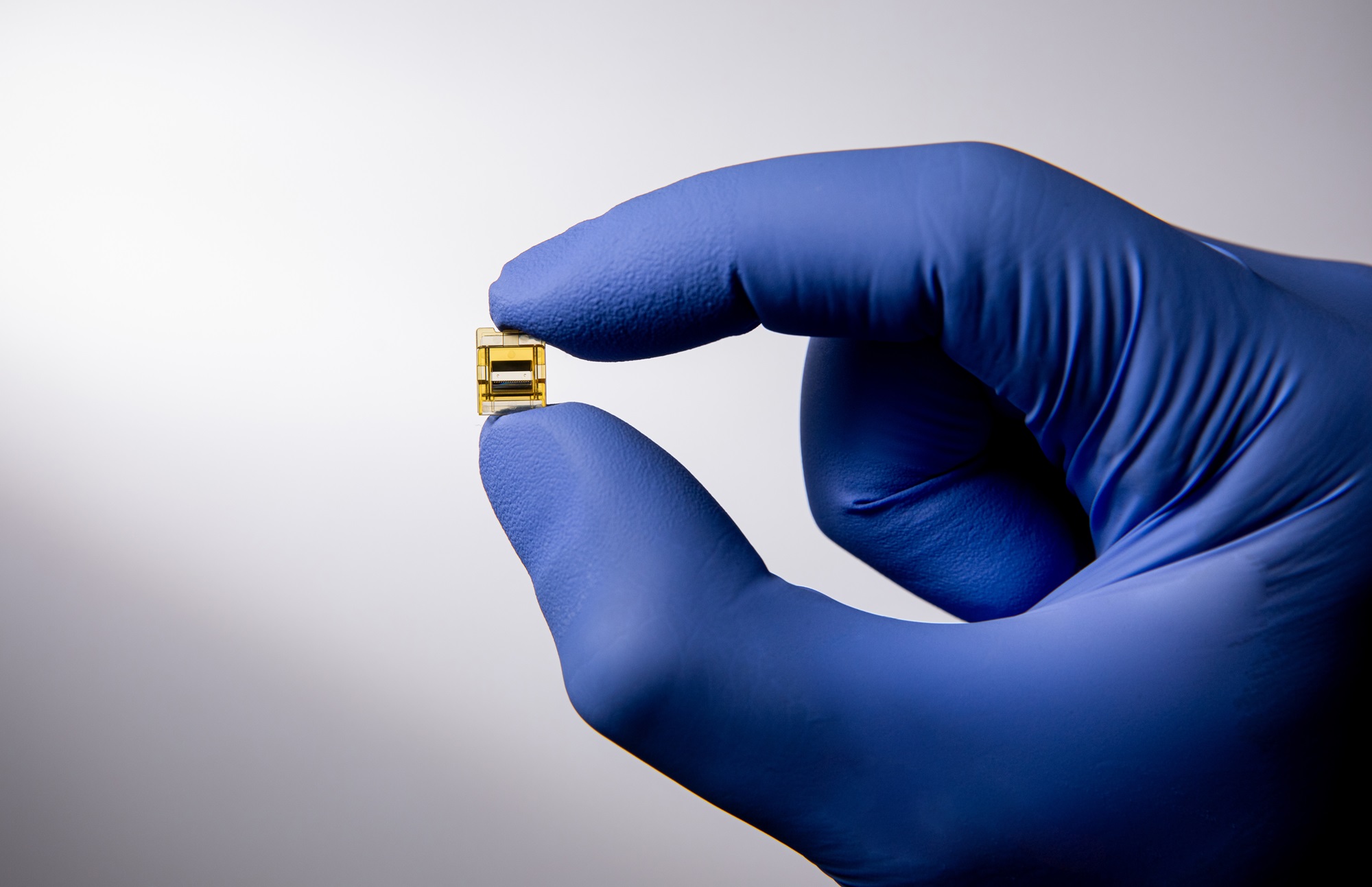
Maximize Your Plastics’ Optic, Mechanic, or Fluidic Performance
Enjoy advanced engineering solutions including design consultancy, performance evaluation, and analytical problem solving with Enplas’s specialty custom productions.
Total Engineering and Manufacturing Solutions
Customers rely on Enplas for engineering, prototyping, and mass production of plastic devices and parts to achieve exceptional standards with a stress-free process.

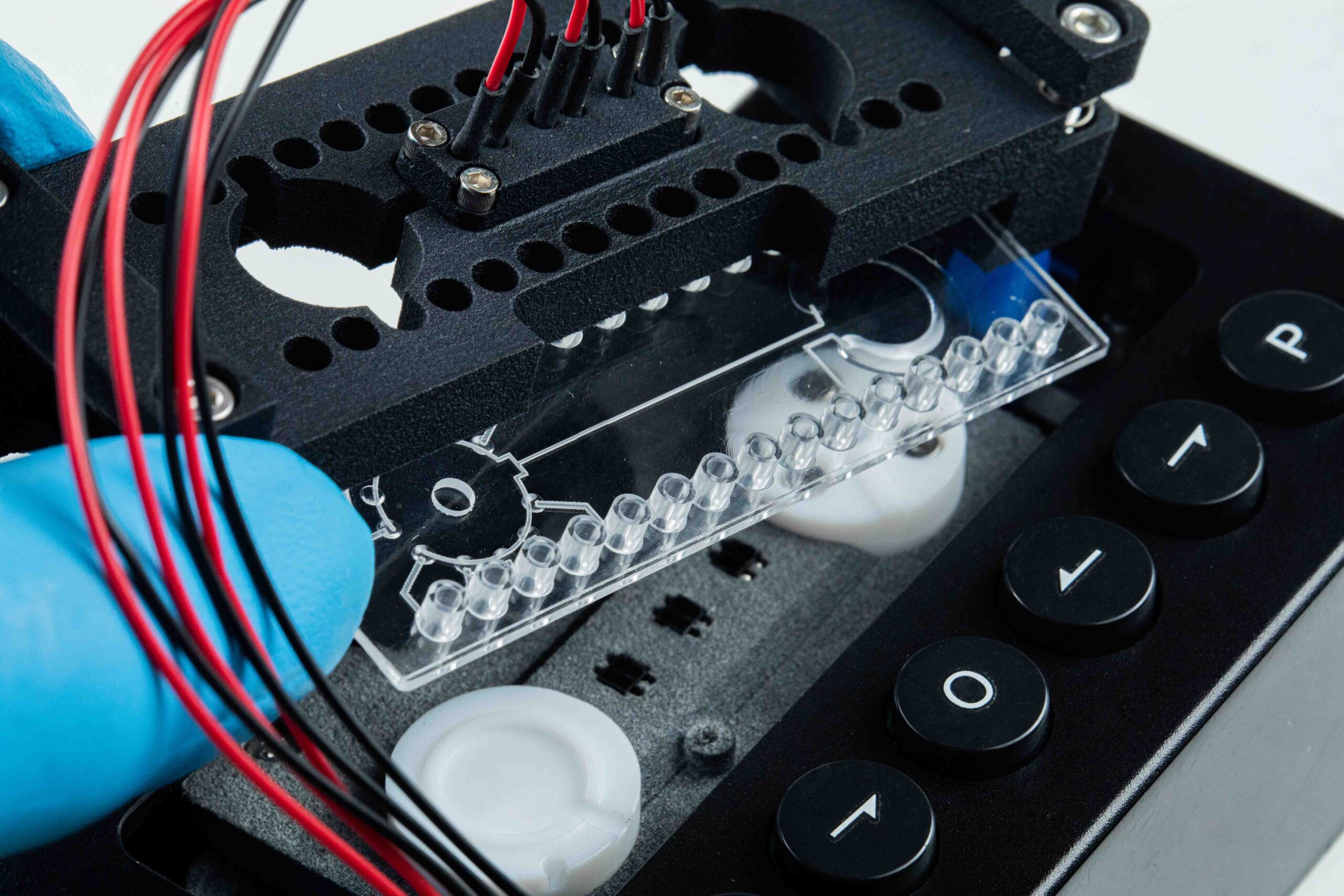
Innovation Lab
Enplas provides innovative and creative ideas to overcome plastic product design roadblocks and enhance device functionality.
News & Highlights
Events
ADLM 2024
Medical & Diagnostic
July 30 - August 1, 2024
McCormick Place Convention Center
2301 S King Drive, Chicago, IL 60616
Booth #3450